- English
- فارسی
A mononuclear PdII complex with Naphcon; crystal structure, experimental and computational studies of the interaction with DNA/BSA and evaluation of anticancer activity
Ramezanpour, A., Karami, K., Kharaziha, M., Zakariazadeh, M., Lipkowski, J., Shahpiri, A., Azizi, N. and Namazian, M., 2021. A mononuclear PdII complex with Naphcon; crystal structure, experimental and computational studies of the interaction with DNA/BSA and evaluation of anticancer activity. Polyhedron, 206, p.115333.
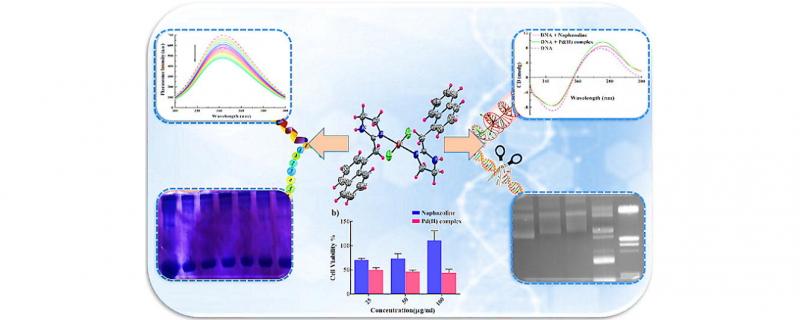
Throughout this study, a new palladium (II) complex, trans-[Pd(Naph)2Cl2], with naphazoline hydrochloride (Naphcon) as an imidazole derivative, 2-(naphthalen-1-ylmethyl)-4,5-dihydro-1H-imidazole;hydrochloride, was synthesized and characterized by elemental analysis, spectroscopic methods (UV–Vis, IR, and 1H NMR) and single crystal X-ray structure analysis. The cytotoxicity of Naphcon and the PdII complex were investigated in vitro against human breast (MCF-7) and cervical epithelial carcinoma (HeLa) cancer cells. The results revealed the higher anticancer activity of complex rather that of Naphcon against MCF-7 cell line. The findings of in vitro studies including fluorescence and UV–Vis spectroscopy, circular dichroism (CD), thermal denaturation and viscosity measurement indicated the interaction of the PdII complex with calf-thymus DNA (CT-DNA) via a combination of covalent and non-covalent interactions, whereas the free Naphcon interacted with CT-DNA mainly through the groove binding mode. Moreover, the ability of Naphcon and the PdII complex to cleave pUC57 plasmid DNA was investigated. In addition, the interaction of both Naphcon and its PdII complex was explored with bovine serum albumin (BSA) by means of absorption and fluorescence spectroscopy. The binding constant (Kb) could be calculated for compounds through these spectroscopic methods. Based on the competitive binding studies using Eosin, Ibuprofen and Digoxin as site markers, the binding site of the PdII complex and Naphcon was found to be located on site-III and I of BSA, respectively. Furthermore, protease activity of compounds was examined under physiological conditions. Finally, to validate all data obtained from biophysical studies, the molecular docking simulation was employed as a computational method.
Journal Papers
Month/Season:
Spring
Year:
2021
